
Meeting the Complexity of Biology by Multiplexing Tech Development
Meeting the Complexity of Biology by Multiplexing Tech Development
Ten years ago, I started my first research project in a science lab. I was tasked to study the effects of a single DNA sequence variant of a single schizophrenia patient. Was this DNA change affecting a cellular function? Could it be causal? My single functional study of this one mutation took two years; it took my lab mentor five years to study four mutations.
As I learned about human genetics, it became apparent that the existing throughput was woefully undermatched to the vastness of human DNA and disease. We could not afford to spend two years per single DNA change. And the problem was even worse for drug discovery. For example, using existing technology, it would take millenia to determine which of the existing 20,000 FDA approved drugs hit any one of our 20,000 human genes. The current approaches were just not acceptable.
Since that first project, I’ve joined teams focused on changing this paradigm. Teams that develop “multiplexed functional assays”: technology to simultaneously test thousands of DNA sequences for their functional effect, for experiments that can be performed in weeks, not years (Starita et al).

The future is finally arriving. Because of the development of multiplexed functional assays, enabled by the last 20 years of advances in massively parallel DNA sequencing and high-throughput programmed DNA synthesis, biology is at a moment similar to the invention of parallelizing processes on a computer. NASA once had to hire 100s of people to make all necessary calculations by hand. Now, a single computer can do the work of billions (or trillions!) of those people.
At Octant, we apply multiplexed functional assays to optimize drug development. In this post, I’ll describe how we use these assays to accelerate their own tech development. In essence, we’ve used our own multiplexed platform to improve itself.
I. USING A MULTIPLEXED ASSAY TO IMPROVE OUR MULTIPLEXED ASSAY
Biology is complicated (citation: our metabolism, cellular development of a 1 mm worm, the mechanics of the SarsCoV2 virus). At Octant, we engineer multiplexed functional assays for GPCR signalling. For us, as in the case of any synthetic biology (synbio) project, the complexity of biology rapidly causes the number of design choices to geometrically expand. Due to biology’s complexity, picking the right one is no easy task. Even after agonizing over a plasmid’s architecture or constitutive promoter choice, scientists can be burned months (or even years) later by an unforeseen experimental “gotcha” (e.g. you missed that you created an accidental feedback loop by choosing a promoter to express your GPCR that itself is activity dependent on GPCR signalling; see Section V for another example).

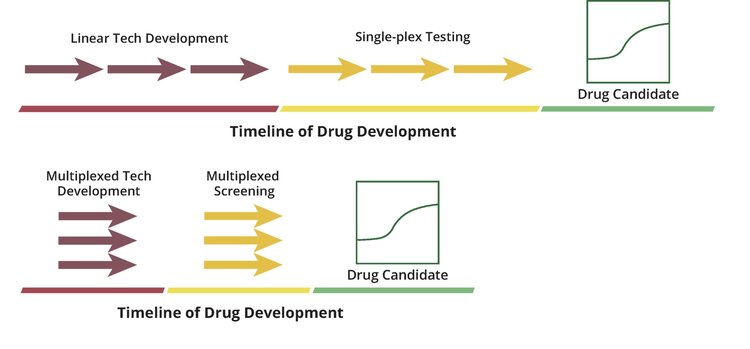
To accelerate our tech development, we use Octant’s platform to rapidly de-risk biological design choices for the plasmids and cell lines that we directly feed back into the platform. Instead of linear iteration, we used a two-step process: 1) simultaneously assay all of our ideas in a single multiplexed experiment (the equivalent of 100-1000s of single-plex experiments), and 2) immediately implement the winners (Fig 1).
This project was motivated by three critical technical design choices (Fig 2 A-C). We used our platform to simultaneously beta-test 80 potential solutions against 20 different conditions (80 x 20 X 4 replicates = 6,400). With this data, and after only 8 weeks of hands-on time, our team created its final design. We dramatically increased the speed of tech development to that required by the drug discovery challenge.
II. NOW, FOR THE SCIENTISTS: SOME EXPERIMENTAL DETAILS AND SOME DATA
I. THE EXPERIMENT
Octant’s screening platform pools up to 100 GPCRs per well in a 384-well dish to screen 1000s of drugs per week. Building off of the massively parallel reporter assay framework development >10 years ago (Patwardhan et al 2009), we use a DNA-encoded barcode expressed from a signalling pathway-responsive reporter to report on each GPCR’s activity (see Naomi Handly’s multiplexing post for detail).
In the early stages of this project, we quickly realized we’d need to optimize three critical components of this technology (Fig 2A):
- the “signalling pathway-responsive reporter” itself (Fig 2A, left)
- constitutive promoters that don’t respond to GPCR signalling (Fig 2A, center);
- a titratable inducible promoter to induce each GPCR’s bespoke amount of expression (Fig 2A, right).
Instead of testing just one idea for each, we decided to multiplex the functional de-risking of the 80 solutions we’d brainstormed. On a single 384 well plate, we tested 80 constructs in a pool against 20 conditions in quadruplicate - the equivalent of 6,400 optimization assays in a single week.

A) The Octant platform required optimization of three different synbio design choices: the signalling-pathway responsive reporter themselves; constitutive promoters that don’t respond to GPCR signalling; and titratable, inducible promoters for GPCR expression.
B) We generated 80 ideas for different solutions to these problems, and cloned them into a barcoded, pooled plasmid library.
C) This plasmid pool was transfected into mammalian cells and D) tested against 20 different conditions on Octant’s platform. RNA/DNA per well was used to measure the expression output of each one in each condition (the equivalent of 1,600 experiments in quadruplicate).
D) tested against 20 different conditions on Octant’s platform. RNA/DNA per well was used to measure the expression output of each one in each condition (the equivalent of 1,600 experiments in quadruplicate).
This experiment had four simple steps:
- Design 80 bespoke sequences that riffed on our three components (Fig 2A);
- Drop each sequence in front of a reporter gene with a unique barcode nestled in the 3’ UTR (Fig 2A-B). (Note for the multiplexing nerds: As these 80 sequences were so diverse, we couldn’t use a single pooled cloning strategy to build and barcode-map, so we built the 80 using about 15 different construction methods. Octant’s OCTOPUS sequencing was ideal for barcode mapping 100s of individual colonies - using OCTOPUS, we cloned, barcode mapped, and whole-plasmid sequenced these 80 constructs in two weeks.);
- Mix these 80 barcoded constructs together (Fig 2B) and transfect them as one into a pool of mammalian cells (Fig 2C).
- Finally, simultaneously evaluate their effect on transcription using a standard multiplexed functional assay metric: comparing the barcode’s RNA representation to its DNA representation (Fig 2D).
In standard multiplexed functional assays, the cell library is usually submitted to one or two functional conditions. However, with Octant’s platform, we were able to test this library against 20 conditions, including: four dose response curves of four different agonists hitting three G-protein pathways, six doxycycline concentrations, forskolin to flood the cell with GPCR secondary messenger cAMP, and ionomycin+PMA to try to flood it with the secondary messenger Ca2+.
III. TUNING INDUCIBLE EXPRESSION
A brief example of the data available from this assay: Each transgenic GPCR requires a unique expression level to properly signal. Usually, we induce this expression by using a TetOn promoter and varying doxycycline (dox) concentrations (Fig 3A). However, on our platform, we are faced with a paradox: we are pooling ~40 GPCRs in a single well that each require a specific dox, but each well can only receive one dox concentration.
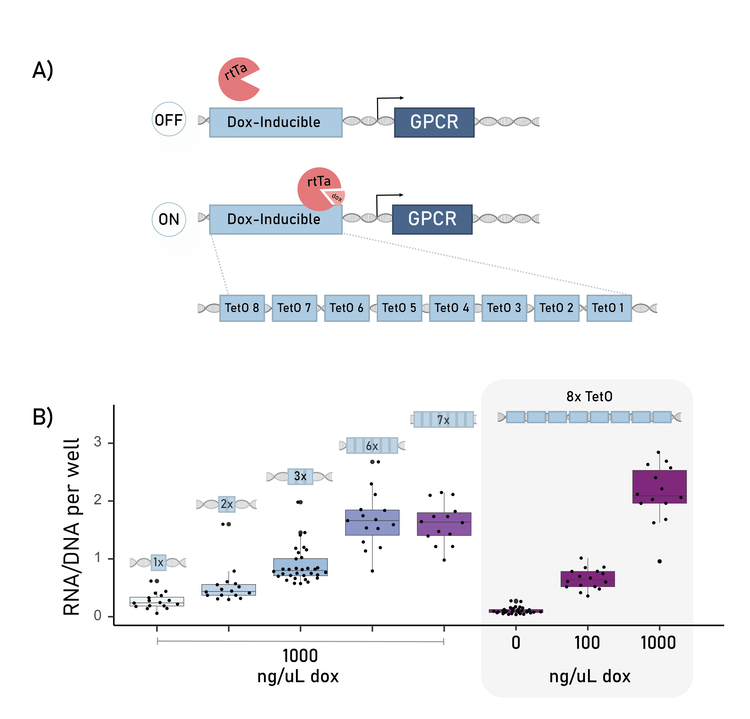
A) The tetOn promoter is an array of 8 tetO binding site motifs. Transcription is induced by doxycycline (dox) treatment.
B) By varying the copy number of the tetO binding sites, the output equivalent of 0 or 100 ng/uL dox treatment can be achieved even with 1000 ng/ul dox treatment.
So, to specify the bespoke inducible-expression for each barcoded GPCRs, we varied the number of transcription factor site motifs in the TetOn promoter. This promoter usually has 8 copies. We tested 1-8 copies of the motif, and demonstrated a clear relationship between copy number and transcriptional output (Fig 3B). From this, we could identified the version that when treated with 1000 ng/ul dox, instead produced the equivalent of a 10 ng/uL dox expression output. This compliments further bespoke-expression tools under development at Octant.
IV. THE FUTURE OF EXPERIMENTS IS MULTIPLEXED
The Octant team is harnessing the power of multiplexed functional assays to not only meet the throughput required by the complexity of biology, but to also accelerate improvement of the technology itself (Fig 1). In drug development, this promises to shorten time-to-molecule and generate richer polypharmacological datasets. We hope these assays will speed solutions for people affected by diseases ranging from cancer to obesity to neuropsychiatric disorders.
If we knew everything about biology, these therapeutic challenges would be easy. Yet we barely have scratched the surface of nature’s vastness. Even though the last 100 years of biotechnology has progressed at breakneck speed (eg. < 80 years since DNA was discovered to be the hereditary molecule; < 90 years since modern computing was first theorized), biology requires light speed. Multiplexed experiments like the one described here can accelerate the arrival of both the tools needed and the solutions for human disease.

ACKNOWLEDGEMENTS
This majority of this project was executed during the 2020 COVID pandemic by an incredible team who contributed to this work in both big and small (but all significant) ways:
Gasperini, Molly. Chan, Henry. Lee, Johnny. Lee-Yang, Grace. Mackenzie, Morgan. Simpkins, Scott. Frederich, Zach. Cooper, Aaron. Haines, Jenna. Jones, Eric. Liu, Winnie. Chan, Leon. Handly, Naomi.
(V. BONUS - BEWARE SV40 SEQUENCES IN HEK293TS: RE-DISCOVERING AN EXPERIMENTAL “GOTCHA”)
IF YOU’RE A SYNBIO NERD LIKE ME (AND MADE IT THIS FAR IN OUR BLOGPOST), HERE IS JUST ONE OF THE “GOTCHAS” FROM WHICH OUR MULTIPLEXED EXPERIMENT SAVED US:
Background: The plasmid library was delivered to cells such that each plasmid should be stably integrated into the genome. The cells were passaged three times to dilute out any transient plasmid. Each construct should have an equal shot of getting in the genome. Thus, when we sequenced the DNA representation of our 80-construct library, we expected to see a roughly equal distribution of each construct.
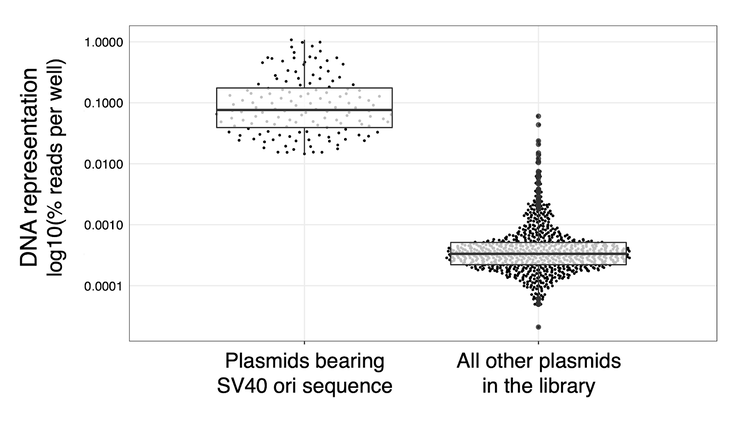
Terror: However, when we first looked at DNA representation, we were horrified: 10 constructs were badly overrepresented (Fig S1)! What could have gone wrong? The 80 plasmids were pooled at equal molarity, they were all transfected into the same cell pool...
A faint bell rang in my head: In grad school, a kind peer had told me a story of how he spent the first summer in lab passaging the workhorse HEK293T cell. For three months, he passaged HEK293Ts to dilute out a transiently transfected GFP-plasmid, but never got the GFP fluorescence to disappear completely. Finally, he presented this stumping result at his first ever lab meeting. A kind soul piped up from the back, “That sucker’s never going away! If you have SimianVirus-40 (SV40) origin of replication on your plasmid, the T-antigen that’s stably integrated into HEK293Ts will replicate the plasmid inside your cells.” (Mahon 2011)
The Gotcha: Staring at this plot 5 years later (Fig S1), I kicked myself that I hadn’t recalled this. All of our overrepresented constructs bore the SV40 minimal promoter, which holds the SV40 origin of replication. These transient plasmids were being continually replicated, wouldn’t dilute out, and had imbalanced our library. We re-ran the experiment without these SV40 plasmids, and successfully achieved a balanced library.
TLDR: Don’t naively use SV40 bearing plasmids in HEK293Ts. This experience underscores the humility with which biology should be approached: even if you’ve learned something once (or twice), it’s easy for most to forget it in the moment. Even if you try to read the literature widely and deeply, it can be so easy for a team to miss something. Plan for the worst data, but hope for the best — and if you can, join a team that can test it all.
Written and Figures by
.png)
Molly

