
Bits and Atoms: My Octant Internship
Bits and Atoms: My Octant Internship
This summer, I was lucky to be Octant’s very first intern! I had a blast. It was a highly unconventional experience, and not just because it happened during the COVID-19 pandemic. I come from a very different background than most Octonauts. Though I’ve taken some formal bio/chemistry courses, I’m an undergrad majoring in Computer Science. My work experience has mostly been in software engineering and AI research.
Why the switch to biotech you might ask? Well, after interning at a big tech company last summer, I did some thinking about what I want in a career. For me, biotech has two big draws: mission and technology. Knowing that your work might one day help people be happier and healthier is a wonderful feeling! Doing so while also solving cutting-edge science and tech problems is a whole other type of thrill. What really sold me on the switch was Octant. After meeting the team and hearing about their unique combination of synthetic biology, chemistry, and computer science, I knew I had found the right place.
Fast forward a few months, and it was time to start my internship. I was assigned to the Chemistry team and introduced to its two members: Galen -- a veteran of the drug industry and the Navy with a love of robotics and high-throughput chemistry -- and Curt -- a polymath scientist who can rattle off facts about everything from yeast metabolism to chemoinformatics.
Chemistry is the newest team at Octant. Curt and Galen came aboard in January and were tasked with a broad mandate: make A LOT of compounds to test on Octant’s drug screening platform. They spent a few months building up a lab to do that. By June, all the pieces were falling into place, and the team was gearing up for lots of synthesis. And by lots, I mean thousands of reactions a week.
What was I doing on this team? In fact, what can a programmer do for a biotech company, especially during COVID-19 while working remotely? Curt and Galen were quite quick at physically performing their 1,000-reaction runs. But post-experiment data analysis was the main bottleneck, eating up most of their time.
Efficient data analysis! That’s a definite software problem, I thought. However, as Galen and Curt dove into specifics, it became clear that plenty of chemistry and science would also be involved.
.jpeg)
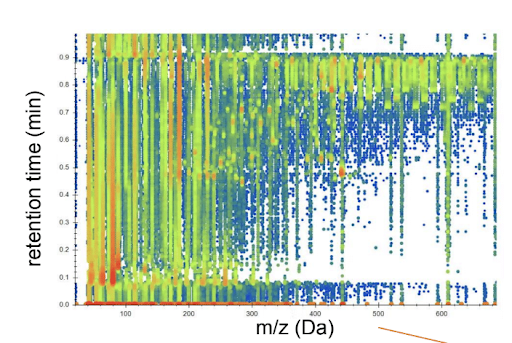
They explained that the Chemistry team wanted to know just a few numbers about each of their reactions: product yield, reactant consumed, and byproducts formed. Unfortunately, no machine could measure those directly. Instead, they fed their reaction mixtures into a liquid chromatograph mass spectrometer (LC-MS). This machine separates a mixture, turns its atoms into ions, and computes each ion’s mass-to-charge-ratio. For one reaction, the LC-MS records 8 million of these mass-to-charge-ratios. Considered together, they hint at the mixture’s composition.
Looking at this sea of data points, Galen and Curt could get a handle of what happened for one reaction using existing software and some manual calculation. Now that they were running thousands of reactions at once, this makeshift method was no longer tenable.
My job, then, was to build a pipeline that would infer each reaction’s molecules of interest (products, intermediates, byproducts) and estimate their presence from our mounds of LC-MS data. The software should also summarize levels of unexpected byproducts and help Curt and Galen identify these mystery chemicals.
Together, we did just that!
The resulting pipeline -- affectionately called Lobster -- predicts products and intermediates using the SMARTS “reaction language.” It then looks for them in our data using their exact mass and isotopic frequency patterns. To unpack this: molecules occur in nature with different numbers of neutrons at calculable probabilities. If a chemical is present in our mixture, we’d expect to see its neutron mass pattern in our data.
.png)
Lobster also compiles a list of features that it doesn’t recognize. It helps us identify these signals by computing their similarity to our reactants. To do this, it takes advantage of fragment data from our LC-MS -- special time points where our machine blasts a molecule into many pieces. Lobster looks for matches between the fragment smithereens of our reactants and the unidentified molecules.
Apart from the raw stats and data-crunching, building Lobster involved plenty of design considerations. It was, after all, a product -- just an internal one. For example, in order to save scientists’ time, Lobster can run across an arbitrary number of worker threads. When executed on Octant's beefy 64-core server, it can plow through a 1,000-reaction data run in 5 minutes. To allow for flexible analysis, Lobster’s main output is a series of tidy dataframes. Scientists can then interrogate the results with their favorite visualization tools: ggplot2 is a household name within the company. Finally, Lobster’s programmatic interfaces allow scientists to easily add stats and visualizations into the pipeline.
In its entirety, Lobster allows Galen and Curt to wade through thousands of reactions at once. They can automatically see which did and didn’t work, while easily noting patterns in successes and diagnosing failures. Notably, Lobster only achieves this by leaning on some wonderful open source packages: OpenMS TOPP and msconvert help us interrogate our data files, RDKIT and brainpy help us predict molecular properties, and Docker helps us package it all together. (And SeeMS from proteowizard helped us make the Pollock figure above.)
I didn’t spend all my time programming, though. After whipping up an early version of Lobster as a remote intern, I worked at Octant in person for the last month of my internship. I was happily given a crash-course in lab chemistry by Galen and Curt and got to perform some really fun experiments. Over a couple of weeks, I even helped onboard some completely new chemistries onto Octant’s platform.

I had a wonderful time at Octant this summer. It’s a company filled with smart, kind people working on fun, difficult, and important problems. Octant is currently seeking its first full-time software engineer. If you’re passionate about the intersection between computation and hard science, please apply!
Posted by

